How do immune effectors influence the structure of the gut microbiome?
Our Research Focus
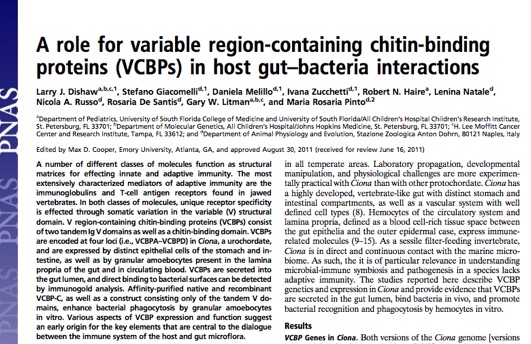
The gut immune system of animals is involved in a continuous dialogue with microbiota as well as diverse antigenic sources; this is a critical interface in the evolution of immune recognition and effector functions. Our research focus is centered on defining conserved phylogenetic signals revealing how host immunity and exogenous factors converge within the gut ecosystem to shape the microbiome. Since most of these trans-kingdom interactions are non-threatening, a primary role for host effectors is likely to help shape interactions among microbiota (the local ecology) and modulate the dialogue between microbes and host interfaces. Furthermore, we are interested in how loss of homeostasis at the mucosal surface of the gut relates to dysbiosis and chronic health problems, such as the onset of Inflammatory Bowel Disease (IBD)-like pathologies observed in humans.
Our lab is also collaborating on additional projects (some with the College of Nursing) that include studies on the developmental maturation of microbiomes in humans, specifically focused on premature birth and development.
Additional collaborations involve projects studying how transkingdom dynamics shape virulence among pathobionts of the human microbiome. Stay tuned.
Additional collaborations involve projects studying how transkingdom dynamics shape virulence among pathobionts of the human microbiome. Stay tuned.
Secreted immune effectors e.g.,VCBPs
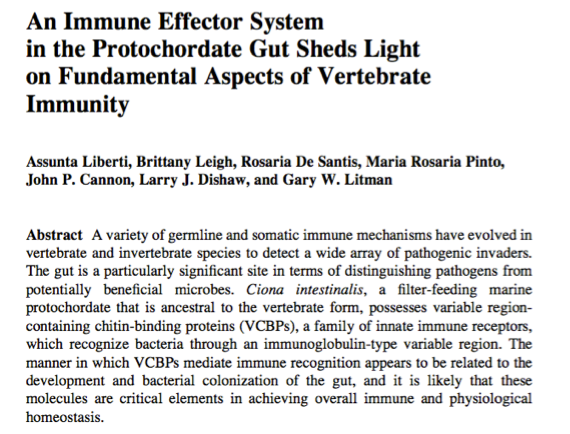
The Ciona gut is colonized by distinct microbial communities; this microbiome is influenced by many factors including environment (biophysics of lumen compartment, bacteriophage, as well as by bacteria "passing through") and host immunity. One host immune protein that likely serves important roles are the Variable Region Containing Chitin Binding Proteins, or VCBPs. We've shown that at least one of these proteins, VCBP-C, is expressed in high amounts in the gut, and can bind bacteria and increase phagocytosis rates (i.e., opsonization) by blood amoebocytes. We have demonstrated that the gut mucus of Ciona is chitin-rich and the VCBPs that are released into the gut are bound by the chitin-rich mucus where they then can interact with bacteria. In vitro, we also show that VCPBs can influence the formation of biofilms. When not tethered to the chitin-rich mucus, VCBP-C can also bind fungi. Thus, VCBPs appear to play an important role in modulating bacterial settlement in the gut, and likely shape interkingdom dynamics.
BACTERIA
While Ciona siphons water 24/7, it can maintain distinct bacterial communities in its gut. We have been able to get many of these bacteria into culture; these isolates have become important study models for us both in vitro and for feeding germ-free animals that we culture in the lab.
BIOFILMS
Many of the bacterial isolates coming from the gut of starved Ciona grow very well as biofilms. This should not be too surprising considering that this is likely their 'normal' lifestyle in the gut. We are utilizing a variety of biofilm-based assays to study these isolates.
PHAGES
Our lab is characterizing the role(s) that bacteriophages play in shaping the composition of host-associated microbiomes. We are also focusing on the role of prophages in the gut of Ciona.
Our work involves both in vivo and in vitro experiments and is supported by the NSF.
Some of our phage work involves a collaboration with Dr. Mya Breitbart.
Our work involves both in vivo and in vitro experiments and is supported by the NSF.
Some of our phage work involves a collaboration with Dr. Mya Breitbart.
VCBPs
Invertebrate chordates like Ciona possess a special immune-type molecule that is expressed almost exclusively in the gut. These variable region containing chitin-binding proteins (VCBPs) are Ig-like proteins that can bind bacteria, increase phagocytosis, and influence biofilms. They are a major aspect of our host-microbiome studies. These secreted immune effectors likely serve critical roles in shaping the ecology of the gut microbiome.
FUNGI
What role(s) do fungi play in the gut? Why are they important?
We are now investigating fungi as an important player in the maintenance of gut microbiome homeostasis. VCBP-C can bind both bacteria and fungi, and may shape interaction outcomes.
We are now investigating fungi as an important player in the maintenance of gut microbiome homeostasis. VCBP-C can bind both bacteria and fungi, and may shape interaction outcomes.
In vitro
We are developing a variety of in vitro assays to mimic conditions of the gut in our studies. Further, we are using 3D matrices to develop 'competition' assays. These 'microbial ecology' experiments are highly relevant to what is happening at the surface of the gut epithelium in Ciona.
Larry
Speaking to undergrads at St Pete College
Britt
Presenting at Gordon Research Conference
Lexi
Presenting at USF Research Day
Annual Workshop
Students from St. Petersburg College
*This website is not an official University of South Florida website. Only the Dishaw Lab is responsible for its content.
Evolution of immune defenses, especially in regards to the gut.
Relationship between symbiosis and immunity
Host-microbe dialogue via innate receptors
The role(s) of adherent bacterial communities in the gut
Molecular phylogeny of immune response genes
Relationship between symbiosis and immunity
Host-microbe dialogue via innate receptors
The role(s) of adherent bacterial communities in the gut
Molecular phylogeny of immune response genes
Our lab maintains active collaborative projects with faculty in various departments and at the All Children's Hospital Johns Hopkins Medicine Campus in St. Petersburg. These are all pilot, IRB approved, studies that focus on various aspects of the microbiome.
Our research is supported by:
National Science Foundation and the
National Institutes of Health
All Children’s Hospital Johns Hopkins Medicine Foundation (several studies)
University of South Florida College of Medicine Deans Office (several early studies in Ciona)